On the Forefront of Cancer Genomics
For more than 40 years, cancer has been the leading cause of death in Japan. Nearly one in two Japanese people is diagnosed with cancer at some point in their lives, and one in four dies of cancer. However, development of new drugs, including molecularly targeted drugs, has improved the 5-year survival rate of patients with cancer, to exceed 60%. Although the outcome depends on the organs, overall, cancer has become a “curable disease.”
Moreover, “cancer genomic medicine (CGM)” can now analyze cancer at the gene level and help select the optimal therapy, and this advancement has garnered significant attention recently. The Graduate School of Frontier Sciences (GSFS) conducts essential “cancer genomics” studies for CGM, with the Department of Computational Biology and Medical Sciences playing a key research role and collaborating with other research institutions outside the school. Sosei invited experts in relevant fields and organized roundtable talks to explore latest developments in cancer genomics at the GSFS.
Session 1:Approach Blood Cancer from the GenomeSession 2:Contribution of Data Analysis to Cancer Genomics
Session 3:Ethics and Economics of Cancer Genomics and Medicine
Development of Cancer Genomics and Cancer Genomic Therapy
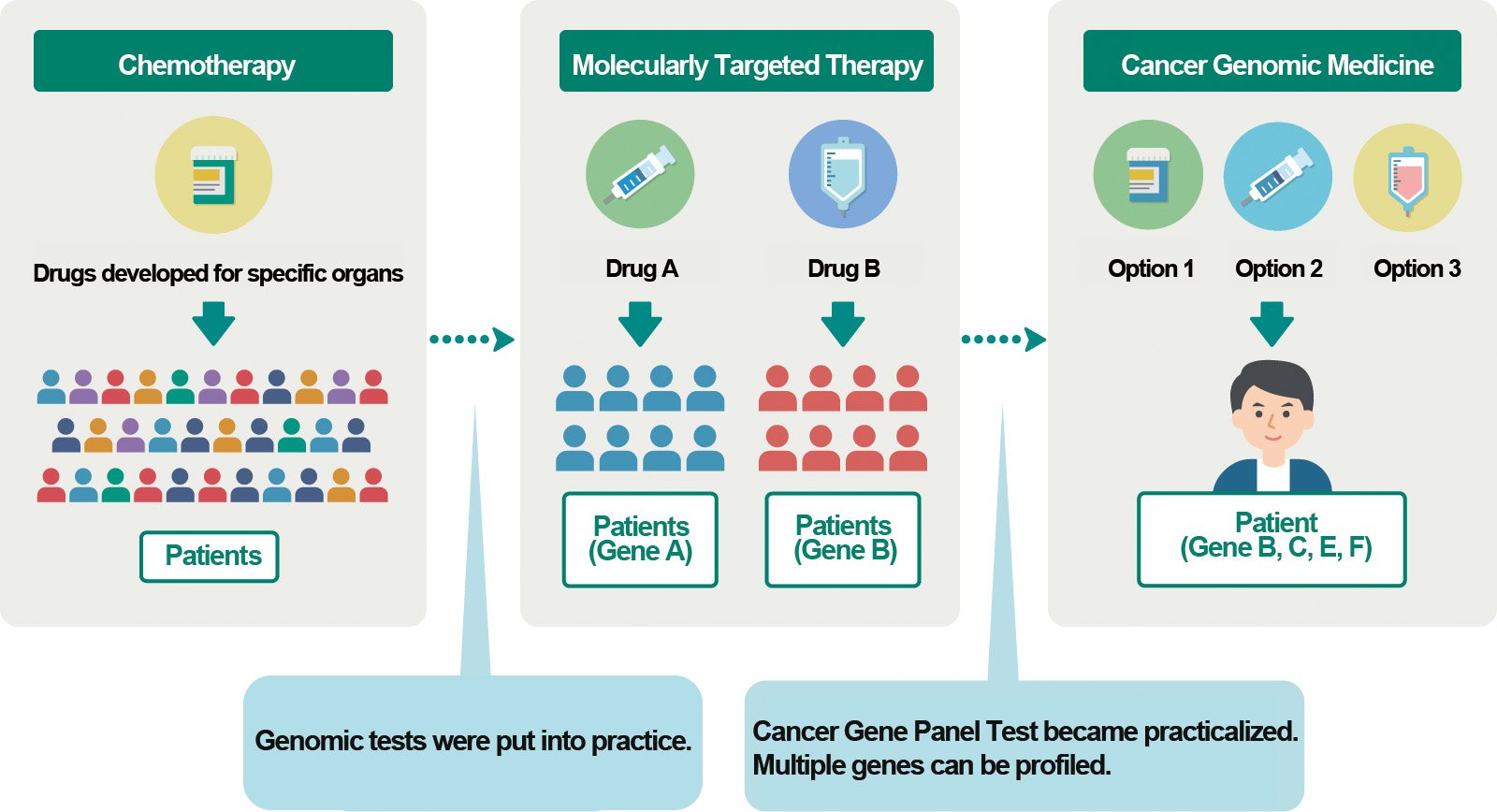
NOTE
The original version of this figure is from the Center for Cancer Genomics and Advanced Therapeutics (C-CAT) website “Simple Cancer Genomics and C-CAT.” The GSFS translated it from Japanese to English with the permission of the author.
https://for-patients.c-cat.ncc.go.jp/knowledge/cancer_genomic_medicine/about.html
Based on the Cancer Control Act, the Japanese government established the 4th Basic Plan to Promote Cancer Control Programs and the 10-Year Strategy of Cancer Research (5th Term) in 2023, with three pillars: “prevent cancer,” “treat cancer,” and “live with cancer.”
In CGM, altered genes are detected by analyzing individual patients’ cancer cell genomes, and the characteristics of the cancer are studied to select the optimal treatment. One cancer gene panel test can detect abnormalities in tens to hundreds of genes. In 2019, the test became covered by insurance for solid cancer, and in 2025, it is also expected to be covered for blood cancer. Thus, the test is almost in the phase of practical application.
However, cancer gene panel testing is available only in designated hospitals, and some conditions must be fulfilled to undergo testing. Consequently, only 10%–20% of patients with solid cancer can receive treatment based on test results. In other words, CGM has just begun. However, various approaches to cancer genomics are imperative to improve the efficacy, accuracy, and range of applications of CGM. The research themes of cancer genomics include whole-genome sequencing, epigenome and multiomics analysis, and the use of big data stored at organizations, including biobank and others. Conducting cancer genomics also requires ethical considerations, including patients’ personal information and awareness of the medical economy. Overall, advances in cancer genomics have paved the way for the development of CGM.
NOTES
Cancer and Genomes:
The human body comprises approximately 37 trillion cells, and DNA of approximately 3 billion base pairs is stored in the nucleus of each cell as a genome. More than 20 thousand genes occupy approximately 2% of a genome, and the genome’s remaining regions are related to gene expression control including epigenomic mechanisms or are regions of unknown function. Many types of cancer develop when multiple abnormalities of genes accumulate in DNA (genome) due to acquired factors, such as errors during cell division, smoking, lifestyle, and aging. However, some are considered to be related to genetic factors. The prevalence of high-throughput machines called next-generation sequencing (NGS) allows the analysis of certain genes or whole genomes and epigenome.
Molecularly Targeted Therapy:
Molecularly targeted therapy is an anticancer drug that specifically targets genes or gene products (e.g., proteins) related to cancer. The development of this type of drug has been increasingly promoted since around 2000. Although therapeutic effects are higher than those of conventional cytotoxic anticancer drugs, critical side effects may occur, and costs tend to be high.
Cancer Gene Panel Test:
It is a method for detecting multiple genomic abnormalities in a patient’s tumor tissues at once to administer the optimal molecularly targeted drug. Since 2019, the test for solid cancer has been covered by insurance, and in 2025, the test for blood cancer is expected to be covered. The test costs 560,000 yen for solid cancer (a patient can undergo the test only once). If the insurance covers 30% of the test cost, the patient can pay 168,000 yen. In Japan, approximately 20,000 tests are conducted annually.
Session 1
Approach Blood Cancer from the Genome
Susumu Goyama
Professor, Department of Computational Biology and Medical Sciences, Facilitator of Session 1 and supervisor of the Sosei Vol. 45 cover story
Makoto Yamagishi
Associate Professor, Department of Computational Biology and Medical Sciences
Yasuhito Nannya
Concurrent professor, the Institute of Medical Science

Goyama: When I started genome analysis about 20 years ago, it was only one of the many fields of research. It has advanced remarkably in the past decade. Finally, cancer gene panel testing for blood cancer is expected to be covered by insurance in 2025.
Nannya: The development of cancer gene panel tests for blood cancer is 5 years behind that for solid cancers, and the target types and cancer stages remain to be determined. However, it will definitely benefit more patients. Additionally, the altered genes often differ at the primary onset and recurrence; thus, the cancer gene panel test should be administered multiple times. However, health insurance would cover the cancer gene panel test only once per patient. In this respect, evidence can be collected through sequencing performed in the research, and we need to show patients that genome analysis has more potential by promoting more research.
Goyama: Professor Yamagishi, how do you use genome analysis in your adult T-cell leukemia (ATL) research?
Yamagishi: In ATL, a human is infected with human T-cell leukemia virus type 1 (HTLV-1). Genomic abnormalities accumulate in the host’s T-cells over 50 to 60 years, and the host develops T-cell lymphoma, which is followed by an extremely poor prognosis. Studying the virus’s characteristics and observing the events occurring in the host are crucial. Once we analyze patients’ genomes, we learn that there are various types of ATL. We analyze patients’ genomes at different times, thus enabling us to predict the prognosis of certain types of genomic mutations or whether transplantation or chemotherapy will be effective for the patient. Therefore, we can use genome analysis to select therapies and predict their clinical outcomes. If a cancer gene panel test becomes common, more data will be collected. I presume that methods for using genome information in clinical practice will be the key to the next phase.
Goyama: I suppose “personalized medicine” based on genomic abnormalities will be developed because we can use genome information more than ever, and we have identified genome abnormalities responsible for different types of cancer. However, if medicine develops in this way, the conventional randomized mega study, which has been the standard in the medical world, will be challenging to conduct. Randomization requires a certain number of samples. Moreover, genome abnormalities and optimal therapies differ among patients. The selection of the best drug for patients based on mega-study results may not be suitable for personalized medicine.
Nannya: Regarding drug effectiveness, biomarkers of molecularly targeted drugs have been simultaneously identified, and patients whose conditions match specific drugs are stratified. However, research has not progressed sufficiently to change standard therapy*1. Therefore, we are developing a system to screen drug effects using the primary cells of patients as part of a joint research project. Every patient has different susceptibilities to drugs, even if they have the same type of cancers. In addition, hematopoietic stem cells and bulk cell populations provide different results. The drugs that we primarily examine for susceptibilities are epigenome drugs*2. Although proposal of effective drugs according to genome abnormalities is often difficult, we continue researching, and hopefully, we can propose optimal drug based on the result of drug susceptibilities.
NOTES
1. Standard therapy:
The best therapy currently available, proven based on scientific justification, and recommended for patients with specific conditions.
https://ganjoho.jp/public/qa_links/dictionary/dic01/modal/hyojunchiryo.html
2. Epigenome:
A collection of states related to the control of genetic expression, including DNA methylation and chemical modification of histones (proteins wrapped around by DNA). The epigenome is passed down through cell division without altering the DNA base sequence.
Goyama: That is an amazing research. In addition to cancer therapies based on genomic information, a considerable amount of cancer epigenome information has been collected. I assume that not only the genome sequence, as the primary sequence information, but also epigenomes, which are related to chromatin structures (collections of histones), will be significant subjects for analysis. Professor Yamagishi, is the epigenome also important for ATL?
Yamagishi: DNA primary sequences and their structures in three dimensions, the genes used, and the time of their use are coded in the epigenome. The epigenomes of patients with malignant lymphoma, such as ATL, differ from those of normal cells. We searched for causes, and it is almost clear that DNA methylation and histone modification are the ones. Unlike DNA primary sequences, the most significant characteristic of epigenomes is their extreme reversibility. If the enzyme that causes epigenome abnormalities is identified, hypothetically, the abnormalities can return to normal conditions. Thus, CGM can be a practical approach for patients.
Goyama: Clearly, the evaluation of epigenomes and primary genome sequences is now necessary. In the meantime, analyzing epigenomes is a challenge because they have immense regions to analyze, are reversible, and alter themselves.
Yamagishi: Our approach is to study gene expression and the epigenomes responsible for them by periodically checking the same patients and observing dynamic abnormalities. In addition, in recent years, a method called single-cell analysis has made it possible to analyze gene expression and chromatin structure in individual cells. This new technology enables us to identify common characteristics of epigenomes in diverse cell populations and epigenomes altered under drug-resistant conditions in a cell population. I believe that one solution to this difficult research is to use innovative technologies skillfully.
Nannya:Valemetostat*3, developed at Professor Yamagishi’s laboratory, is now administered in clinical settings. This is a significant achievement.
NOTE
3.Valemetostat:
It is a novel drug, an inhibitor of methylated histones, developed in Japan. Yamagishi’s laboratory and Daiichi Sankyo Co., Ltd. developed a compound that inhibits the activities of the enzymes EZH1 and EZH2, which induce epigenetic abnormalities found in many types of cancer. Valemetostat normalizes gene expression patterns of ALT and malignant lymphoma, for which effective treatment has not been developed, and it pinpoints cancer cells and kills them.
Goyama: I used to work as a hematologist. Now, I concentrate on basic research, developing new therapies against hematopoietic organ tumors. Over time, genome analysis and genome editing technologies, such as CRISPR, have been developed, and candidate molecules that can be targets for cancer therapy have been increasingly identified. On the other hand, except for a few successful examples, such as Professor Yamagishi’s Velemetostat, developing novel drugs remains difficult even if the causes are identified. In other words, therapeutic development has not advanced substantially. We aim to connect our basic research outcomes directly to clinical medicine by developing nucleic acid drugs that directly target genomes and RNA as digital information. My main goal is to develop innovative cancer therapy.
Thank you for your participation in the discussion today.
Session 2
Contribution of Data Analysis to Cancer Genomics
Yutaka Suzuki
Concurrent Professor, Director of Life Science Data Research Center, Facilitator of Session 2 and supervisor of the Sosei Vol. 45 cover story
Katsuya Tsuchihara
Visiting Professor and Director of the Exploratory Oncology Research and Clinical Trial Center, National Cancer Center Japan
Ayako Suzuki
Associate Professor, Department of Computational Biology and Medical Sciences

Y. Suzuki: Since the installation of Japan’s first “next-generation sequencer” at the University of Tokyo’s Kashiwa Campus in the mid-2000s, comprehensive cancer genome analysis has become possible. Around the time the sequencer was installed, I first met you, Professor Tsuchihara and Professor Ayako Suzuki.
Tsuchihara: It was also when molecularly targeted drugs appeared, but it took a decade for molecularly targeted drugs and next-generation sequencing (NGS) to integrate effectively. In 2019, comprehensive genome profiling tests, as known as cancer gene panel tests, became covered by health insurance in Japan. However, it should be noted that Japan has many types of genomic tests in addition to panel tests. When we talk about CGM, we need to include them. Moreover, for both research and human resource development, concentrating only on genomes is not appropriate, and working with people at clinical sites and in the pharmaceutical industry is crucial. Therefore, since beginning our research, we have collaborated to become Japan’s starting point for genomics.
A. Suzuki: I am working on visualizing how molecules alter according to interactions between cancer cells and immunocytes in the microenvironment using new technologies, such as long-read sequencing and spatial transcriptome analysis. Clinicians tell me that seeing visualized data is interesting because it clarifies what they have vaguely estimated. However, if you ask me whether I have discovered anything, I will answer “no.” I would like to contribute directly to clinical sites by analyzing the problems in depth and acting as a coordinator for those in the basic research field and on clinical sites.
Tsuchihara: I understand what technical researchers consider problems, but I have a different perspective. In the clinical application of genomics, we question why a drug is effective or ineffective and what happens when it becomes ineffective. What the pharmaceutical industry calls “mode of action” and “proof of concept” are what they expect from genome and omics analyses. The clinical site’s top request is thorough analysis for acquiring data to understand what they treat.
Y.Suzuki: In cancer research, the cycle of technological revolution is constantly accelerating. It seems better to leave clinical sample analyses to pathologists. As basic scientists, we would better concentrate on in vitro experiments and observe changes in cell culture, such as “organoid”*1, in real time or approach responses under different conditions.
NOTE
Organoid: A miniature organ produced from stem cell(s) in vitro. Stem cells can self-organize into a three-dimensional organ-like structure through their self-renewal and differentiation capacities.
Tsuchihara: The foundation of genomic analysis has been established, and practical applications have been promoted during the past decade. Here is my question. Is it a bad thing to have mutations in the genes responsible for cancer? The time at which genomes are most intact is when we are born. If we consider such a state “healthy,” do we ultimately have to return to that birth state? I do not want that. I am not talking about genomics. It is a question of the philosophy of life. I enjoy my life better than before, having gone through many experiences. This is also a question about the meaning of cancer therapy. I hope that young researchers have this perspective. Keeping this question in mind will definitely give them a unique idea for their research.
Y.Suzuki: My recent concern is the decline in the researcher population. Academia’s mission is to develop human resources with a broad spectrum of perspectives. To achieve this, grassroots networking among researchers is crucial. The Department of Computational Biology and Medical Sciences produces various types of graduates, and I will continue to expand the network.
Session 3
Ethics and Economics of Cancer Genomics and Medicine
Kaori Muto
Concurrent Professor, Institute of Medical Science
Eiko Saito
Associate Professor, Sustainable Society Design Center,
Yoichiro Kamatani
Professor, Department of Computational Biology and Medical Sciences, Facilitator of Session 3 and supervisor of the Sosei Vol. 45 cover story

Kamatani: In cancer genomics, societal viewpoints, including ethical and economic matters, are very important. First, careful consideration is necessary when obtaining patients’ genetic information, namely, genome sequences. I would like to ask you about the problems or challenges that clinical sites face with these perspectives.
Muto: My specialty is medical sociology, with a particular focus on the ethical, legal, and social issues (ELSI) surrounding genomics and genomic medicine. For about 20 years, I have studied how patients and the public perceive cancer genomics and related medical practices. Cancer patients often express a strong desire for research to advance and for their data to be used as much as possible. Many say they hope future patients will benefit, even if it is too late for them personally. For example, genomic data is stored at the Center for Cancer Genomics and Advanced Therapeutics (C-CAT)*1 in Tsukiji, Tokyo. We asked patients who had undergone a cancer gene panel test whether they would permit third parties, including private companies, to use their data. Approximately 98% of respondents agreed to such secondary use. I believe it is necessary to maintain respectful relationships with patients and to never take their generous intentions for granted.
NOTE
1. The Center for Cancer Genomics and Advanced Therapeutics (C-CAT):
It was established as part of the National Cancer Center Japan. Approximately 20,000 cancer gene panel tests (comprehensive genomic profiling) are conducted annually under health insurance at over 200 medical institutions, including major, base and cooperative hospitals for CGM. The center collects clinical and genomic information from those who can undergo the test and agree to the use of their data.
Patients are generally supportive of the use of their data if it is beneficial for the advancement of medicine, but they may also be wary of potential social disadvantages, particularly if their cancer is hereditary. Meanwhile, when genetic testing indicates a predisposition to cancer, healthcare providers believe that patients should inform their relatives, including their children, about the possibility of carrying the same genetic traits. Definitely, this is a crucial first step for cancer prevention.
However, an interview study of patients with hereditary breast and ovarian cancer syndrome and their families revealed that some patients experienced difficulties with their relatives. Some felt guilty when their relatives developed cancer after they had hesitated to share the information. Certain types of cancer are hard to detect in an early stage or require consultations across multiple medical departments. Some patients said that the efforts to share information about hereditary cancer risk were not worthwhile. It is unreasonable to place the full burden of communication on patients, not only in the case of cancer but for any hereditary disease. Society needs to raise awareness that genetic alterations are natural phenomena. Creating an environment where people can talk openly about genetic risks, with the support from healthcare providers and patient communities, is important for fully realizing the benefits of genomic medicine.
Kamatani: Prof. Saito, you specialize in health economics. Would you please tell us something from your perspective?
Saito: I began working at the Sustainable Society Design Center, GSFS, in April 2024. Previously, I worked for the National Center for Global Health and Medicine, and before that, for the National Cancer Center Japan, evaluating the health economics of a cancer gene panel testing.
Two perspectives are needed for health economics analysis of cancer gene panel testing. The first is cost-effectiveness, and the second is the impact on the national budget, namely, the influence on national health financing. Simply speaking, cost-effectiveness is the ratio of the cost of intervention for gaining one healthy year. The conditions to determine the cost-effectiveness of a cancer gene panel testing include: a test was conducted appropriately, a molecularly targeted drug that matched the patient’s condition was applied, and the patient recovered well. The problems are that the drug options suggested by the current cancer gene panel tests are limited, and do not always lead to a better prognosis for some types of cancer.
Consequently, the cost-effectiveness of cancer gene panel testing is determined on a case-by-case basis. Moreover, because of the extremely high cost of molecularly targeted drugs and the consequent burden on the national budget, the impact on the health financing must also be evaluated. Therefore, we need to evaluate the nation’s budget impact for cancer gene panel tests conducted nationwide along with cost-effectiveness.
Health economics uses two types of research methods. In recent years, the first method is evaluation using real-world data*2. Health economics specialists join the clinical team from the intervention and clinical trial design phases to determine the cost measurement method and data collection. If an inquiry form asking about patients’ quality of life (QOL) is needed, we include it and measure the outcome index. The advantage of this method is that it allows us to measure the cost that matches the nation’s situation and evaluate a broader range of health economic impact, including targeted patients’ QOL.
The second method is the traditional modeling method. We collect data from previous studies in and outside Japan, such as cost, effectiveness and patients’ QOL, which can have various parameters. By inputting them into models, we can analyze and obtain evaluation results quickly through secondary data use.
If we use meta-analysis, the evidence level becomes more concrete, but we do not know if it fits the targeted patients’ conditions. Both methods have advantages and disadvantages. Real-world data are more precise but have logistical and cost challenges. This also includes an ethical issue regarding patients’ burdens. I personally think the best strategy is to use only each method’s advantages.
NOTE
2. Real-world data:
Information and data obtained at clinical sites or from the real world, such as pharmacy receipt data, insurers’ data, electronic medical records and patient report outcomes
Kamatani: When conducting cancer genome tests, there have been controversies about whether to conduct a single gene or panel test and whether to conduct an exome or whole-genome analysis. If we conduct a whole-genome analysis of three billion bases, we can understand many more things for research. However, whole-genome sequencing is significantly more costly than exome analysis, which is a financial disadvantage. What do you think about this?
Saito: In medical economics, research and clinical services are completely different stories. Research does not always produce results—this is acceptable—but at clinical sites, only effective results obtain investment. We cannot talk about research and clinical services at the same time. In Japan, we cannot say anything about how much economic return can be expected from conducting whole-genome analysis and developing novel drugs because, so far, no data exist.
However, when we need to consider more multifaceted value measures, such as patients’ hopes for therapies and fears of death, medical economics also suggests evaluating how much the cancer gene panel test contributes to science. From this perspective, I believe that whole-genome analysis is significant.
Kamatani: I have been analyzing various genome data, including those from cancer. If researchers and pharmaceutical companies share the genome data of cancer and intractable diseases collected at medical sites, based on the society’s demand, medical services will improve. Hopefully, we can analyze data from such a good cycle.
It was good to know that patients are very cooperative with medical advancements needing ethical consideration. In addition, through today’s discussion, I learned that new research, such as real-world data, is being promoted in medical economics. I hope to contribute to the future of cancer genomics and medicine with researchers like you. Thank you very much.
After Discussions
Through this cover story, I hope that readers will learn that cancer genomics is advancing every day and that CGM is incorporating research outcomes. However, matters remain for exploration and improvement. Moreover, ethical and economic perspectives are essential for developing CGM. I hope that both research and clinical sites will form a harmonious circle.
Interviewed, written and edited by Kazutada Furui
vol.45
- Cover
- On the Forefront of Cancer Genomics
- Visualizing the Universe: Exploration of Plasma in the Planetary Atmosphere and Space
- Finding Biological Information in Genetic Sequences
- Future of a Sustainable Low-Carbon City by a Transdisciplinary Approach
- GSFS Front Runners: Interview with an Entrepreneur
- Voices from International Students
- On Campus/Off Campus
- Events & Topics 1
- Events & Topics 2
- Information
- Relay Essay
