Frontier Sciences
Frith Martin
Finding Biological Information in Genetic Sequences

Genetic sequences hold information for how all living things function and interact. These sequences slowly evolve and retain traces of their history and relationships from billions of years ago. The Frith group performs computational analyses on these sequences to obtain interesting information. We try to retain the feelings of wonder, excitement and play.
Frith Martin
Professor
Laboratory of Large-Scale Bioinformatics
Division of Biosciences
Department of Computational Biology and Medical Sciences
https://sites.google.com/site/frithbioinfo
In one example, we found some DNA sequences that control gene expression were subtly conserved in diverse animals. Figure 1 shows DNA sequences that control expression of the Sp5 gene in humans and animals, including sea urchins, oysters and coral. Amazingly, these sequences have been partly conserved since these animals’ last common ancestor lived—in the Precambrian—about 700 million years ago. Intriguingly, all such conserved sequences were found to control gene expression during embryonic development. This shows “deep homology” of development between animals as different as humans and coral; it reveals a control system for animal development conserved since the Precambrian.

Figure 1
Sp5 Promotor
In another study, we identified genetic “fossils” of viruses and virus-like entities (transposable elements) in the human genome. Sometimes, DNA from these entities is inserted into the host organism’s genome. Figure 2 shows an example: at the top is an amino acid sequence of a retrotransposon protein (reverse transcriptase); below is human DNA and its translation into amino acids. The DNA sequence has “decayed” through mutations occurring over millions of years and retains only subtle similarity to reverse transcriptase. Amazingly, this DNA was inserted into the host genome before the last common ancestor of all jawed vertebrates, about 450 million years ago. We know this from comparing human and shark genomes to see that this part of the genome is conserved. We usually think of fossils as subtle traces in rocks, but they can also be traces in genetic sequences. These genetic fossils are the main method of studying ancient viruses, a field called paleovirology.

Figure 2
Genetic Fossil
Our primary approach to obtaining information from genetic sequences is probability calculations. By comparing related sequences, we can determine the rates of different types of substitutions, insertions and deletions. Then, we can use these rates to determine whether sequences are subtly related.
Sequences can also evolve in more complicated ways, with duplications and complex rearrangements. In such cases, we can also use probability calculations to determine the likely ways sequences are related. We used this method to characterize complex chromosome rearrangements that cause disease in humans.
Using efficient algorithms is also important because sequence datasets are often large. One method is to sample a subset of positions in a sequence. For example, positions where aa occurs or where ac occurs. At first, these appear to be equally good: aa and ac occur equally often in a random DNA sequence. But there is a surprise! In a completely random DNA sequence of length 3, are aa and ac equally likely to occur? The surprising result is shown in Figure 3. This means that the sampling methods are not all equal (ac is better than aa). We can leverage this counterintuitive effect to optimize the efficiency of sampling a sequence.
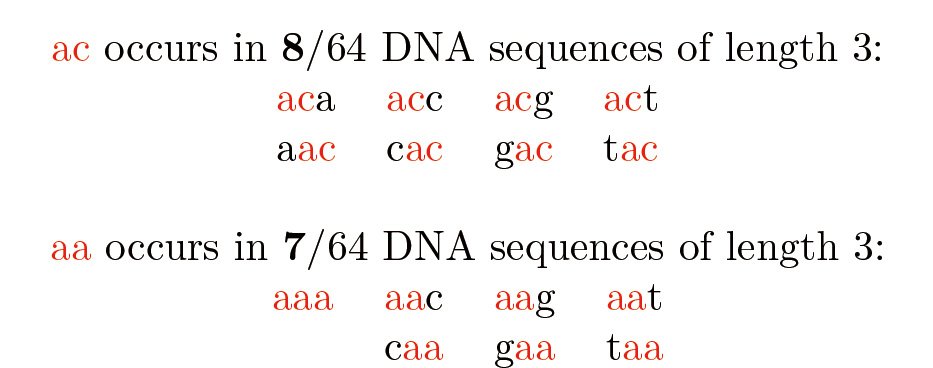
Figure 3
DNA Word Count
vol.45
- Cover
- On the Forefront of Cancer Genomics
- Visualizing the Universe: Exploration of Plasma in the Planetary Atmosphere and Space
- Finding Biological Information in Genetic Sequences
- Future of a Sustainable Low-Carbon City by a Transdisciplinary Approach
- GSFS Front Runners: Interview with an Entrepreneur
- Voices from International Students
- On Campus/Off Campus
- Events & Topics 1
- Events & Topics 2
- Information
- Relay Essay
